RESEARCH INTERESTS
In their lifetime, 1 in 2 men and 1 in 3 women are diagnosed with invasive cancer. The perpetuation of a tumor hinges largely on cancer cell-intrinsic signals maintaining tumor growth and the tumor’s ability to evade destruction by the immune system (Obenauf and Massagué, 2015). These dependencies are exploited by targeted therapies, which inhibit cancer cell-intrinsic signaling pathways, and immunotherapies, which unleash an immune response against cancer cells. Targeted therapies and immunotherapies have shown remarkable success in subgroups of patients, but therapy resistance and low response rates pose daunting challenges (Haas et al., 2018). The combination of different therapies is a promising path to overcome low response rates and acquired resistance. However, rational combination and optimal sequencing of therapies are hampered by a lack of molecular insight into resistance mechanisms and the interplay of therapies.
Our lab studies the molecular determinants of response and resistance to targeted and immunotherapies with the vision and mission to guide rational combination therapies to achieve durable responses in cancer patients. We aim to answer fundamental questions in cancer biology, such as: How is immune evasion facilitated? What contribution have the oncogenic pathways that drive tumor initiation in evading the immune system during metastasis or therapies? How are cancer cells and their microenvironment evolving during the inhibition of oncogenic pathways or the stimulation of an immune response? Which challenges and opportunities arise from this?
We study these general principles in MAPK-driven cancers of different cellular contexts, including melanoma, lung, colorectal, and pancreatic cancer, which represent cancer types with high to low capacity to respond to immunotherapies and are amenable to the treatment with MAPK inhibitors (BRAFi, MEKi, and the newly developed KRASi) (Obenauf et al., 2015). In addition, a sub-group of the lab investigates the molecular drivers rare skin cancers, such as Merkel cell carcinoma (Leiendecker, Jung, et al., 2020). For our studies we use human and murine cell line models, which easily allow the functional perturbation of candidate genes and can be transplanted in vivo to study interactions with the tumor stroma and a functional immune cell compartment. To complement these studies, we use patient derived xenograft models, genetically engineered mouse models, and co-culture systems and work closely with clinicians to cross-validate our findings in patient data. We are developing novel tools (e.g. CaTCH, Umkehrer et al., 2020), perform whole genome CRISPR screens with various readouts, and a whole plethora of biochemical and genetic approaches to uncover novel mechanistic insights.
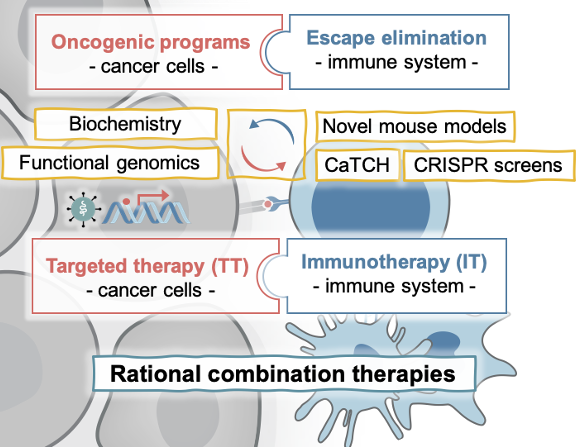
KEY CONCEPTUAL ADVANCEMENTS AND ONGOING PROJECTS
Development and application of a new functional lineage tracing tool using CRISPRa-inducible reporters
In our manuscript published in Nature Biotechnology (Umkehrer et al., 2020), we describe a novel lineage tracing tool termed CaTCH (CRISPRa tracing of clones in heterogeneous cell populations). CaTCH combines precise mapping of the lineage history of millions of cells with the ability to isolate any given clone alive from a complex population based on genetic barcodes. Thus, CaTCH enables the retrospective isolation and analysis of founding clones from heterogeneous cell populations before evolutionary selection with a remarkable resolution of 1:50.000 (0.002%) and purity (98%, ~20,000-fold enrichment).
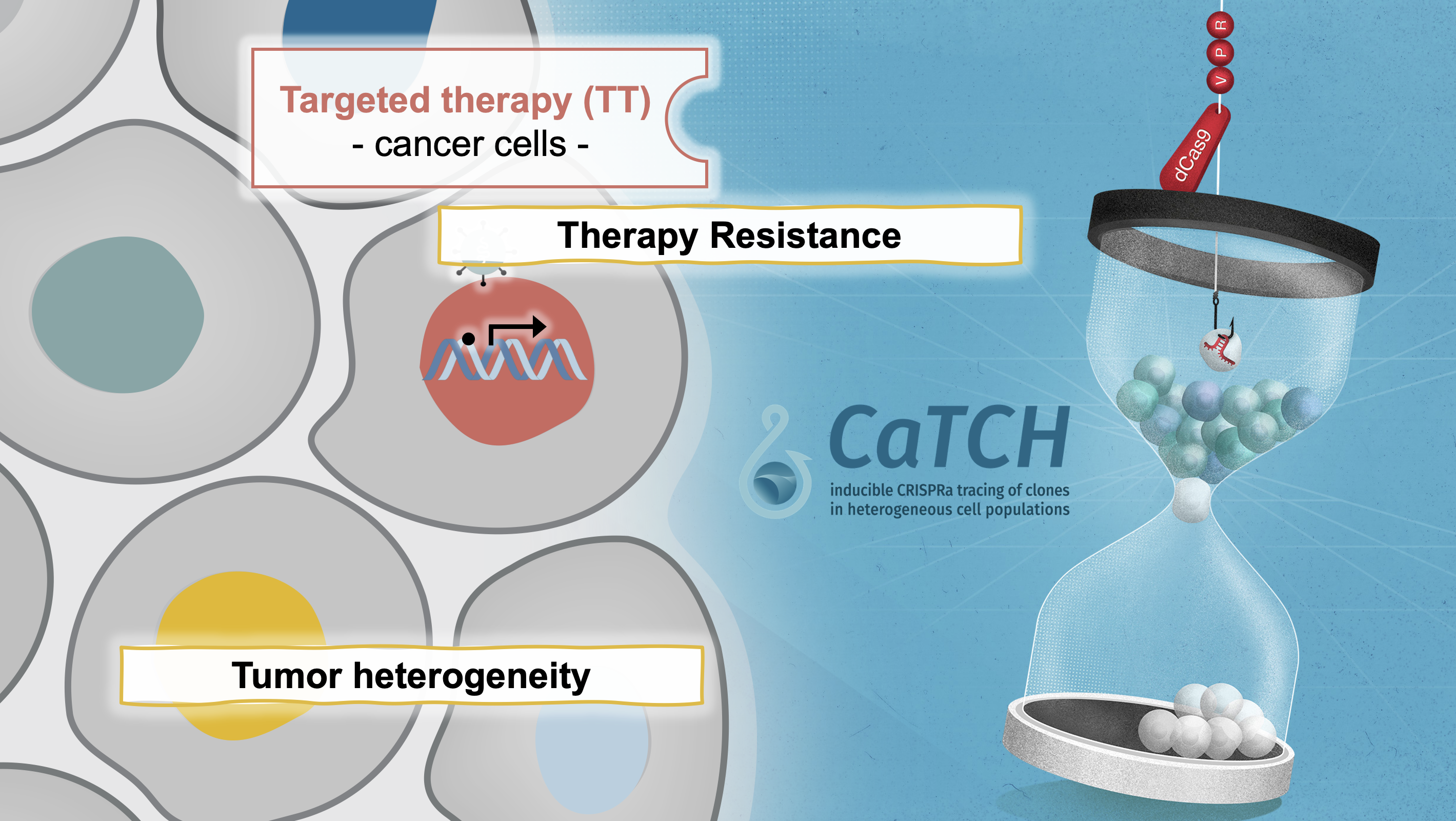
In first applications, we used CaTCH to provide insights into resistance to targeted cancer therapies in vivo. In our model, we find that most clones can acquire resistance to combined RAF/MEK inhibitor therapy, indicating that resistance to this clinically relevant regimen is a universally achievable state. We found a de novo KrasG12R mutation in a resistant clone, but not in its treatment-naïve counterpart. Our results provide experimental evidence that mutations are acquired during drug treatment, which is difficult to prove using prior methods, and challenges the current notion that mutations conferring drug-resistance generally pre-exist before therapy. If you are interested to read more about how we developed CaTCH, you can read our blogpost on the Nature webpage.
While this first report focused on the establishment and validation of CaTCH as an experimental tool in the context of targeted therapy resistance, it opened several new lines of research in our lab allowing us to mechanistically dissect resistance to various therapies, immune evasion, and metastasis. Finally, we envision that CaTCH will allow to address fundamental questions in basic and translational research, e.g., how cell identity states and trajectories are determined in therapy resistance, metastasis formation, tissue development, and somatic cell reprogramming. We are very happy to share the CaTCH technology with anyone that is interested. The basic vectors can be found on Addgene. If you are interested in the analysis tool (including Galaxy integration) as well as the CaTCH barcoding library, please reach out to us directly (anna.obenauf@imp.ac.at).
Mediators of response and resistance to targeted and immunotherapies
Understanding the contribution of oncogenic pathways towards driving disease progression and immune evasion
To better understand the clinical emergence of resistant cells to targeted therapies our work also investigates the poorly understood events during tumor regression. In recent work, we discovered that inhibition of the oncogenic driver pathways using targeted therapy with kinase inhibitors induces a complex network of secreted signals in drug-stressed melanoma and lung adenocarcinoma cells, which ultimately drives the relapse of those tumors (Obenauf et al., 2015). This response, termed therapy-induced secretome (TIS), does not only enhance the survival of drug-sensitive cells, but also stimulates the proliferation, invasion, and metastasis of drug-resistant clones that are lurking in the background of the regressing tumors (Figure 2a-c).
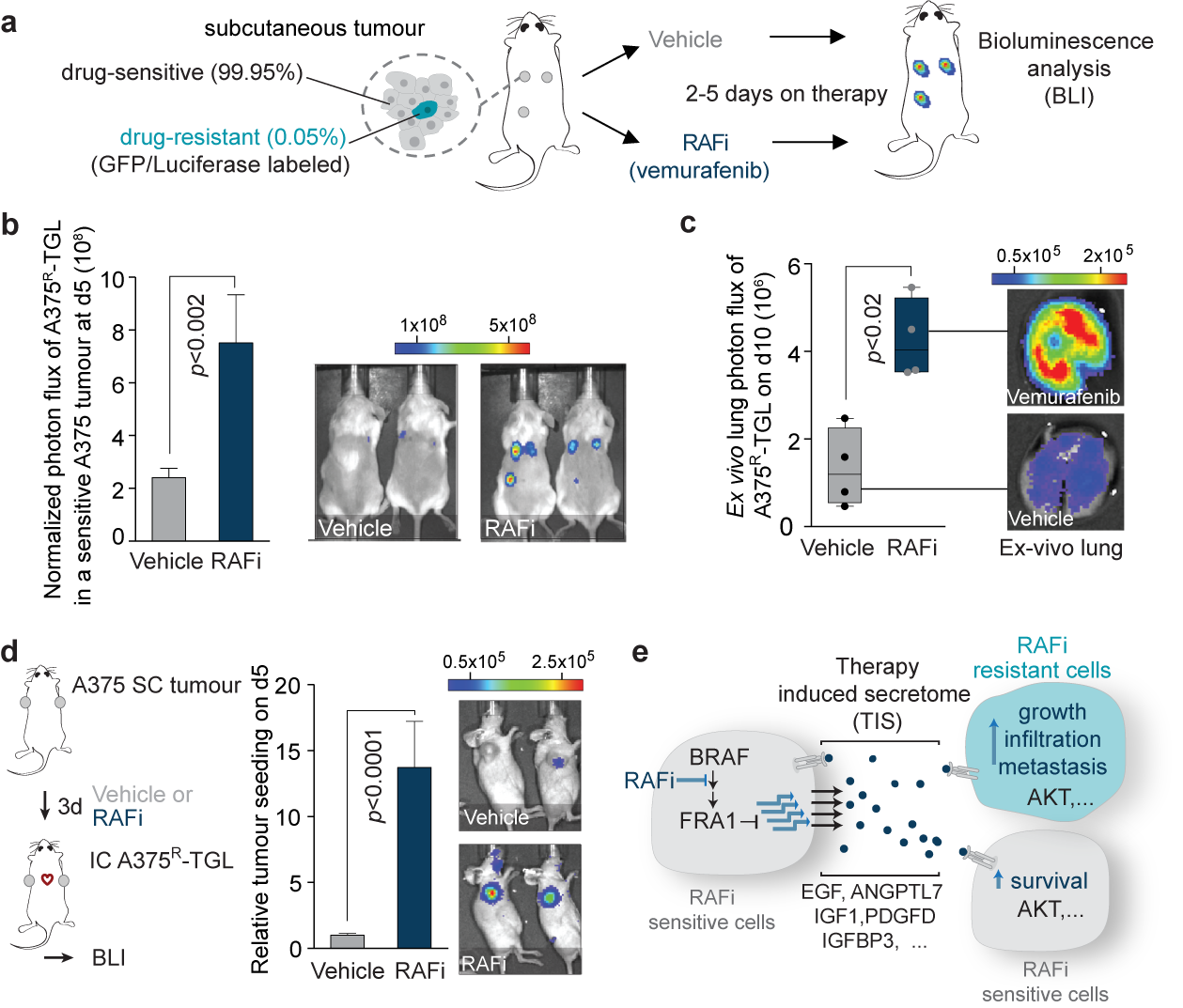
We also found that the regressing tumors in our animal models act as potent ‘magnets’ to attract drug-resistant cells from the circulation. This process, termed self-seeding, could add an additional layer of complexity to the treatment and relapse of patients on targeted therapy (Figure 2d). Our findings establish a general mechanism by which drug-stressed tumor cells can aggravate cancer progression and it could – at least partly – explain why targeted therapies rarely lead to complete tumor regression. By dissecting the processes that lead to this phenotype in melanoma, we identified that the TIS consists of a vast amount of signals that are capable of activating multiple signaling pathways, including an important survival and proliferation pathway (PI3K/AKT/mTOR pathway) (Figure 2e). The addition of AKT/PI3K/mTOR inhibitors blunted the outgrowth and metastasis of resistant cancer cells in animal models. These experiments suggest that this drug combination is a potential strategy to delay tumor relapse in patients. Even more importantly, our study started to expose the significant changes in tumors treated with targeted therapies. These changes are still largely unexplored and are expected to have a major influence on the efficacy of other drugs, such as immunotherapies, when given in combinations.
Cancer therapies are often administered sequentially, with the subsequent lines of therapy being applied after a tumour relapses on the first-line treatment; however, how therapies shape the tumour and thereby influence subsequent therapeutic response is poorly understood. In an ongoing project we focused on BRAF-mutated melanoma, where the current ASCO and ESMO guidelines recommend as first-line treatment either targeted therapy with MAPK pathway inhibitors or immunotherapy with checkpoint inhibitors. However, there is no clear mechanistic understanding which choice of first-line therapy is better. We have closely worked with clinicians to obtain patient data and generated novel mouse models that mimic the different phases of treatment (treatment-naïve tumors, tumors responding to therapy, and tumors resistant to targeted therapies) and made some striking observations. More coming soon…!
Mediators of immune evasion and novel approaches to enhance an immune response
A large fraction of metastatic cancers does not respond to the clinically approved checkpoint inhibitors anti-PD1/anti-CLTA-4. A number of projects in our lab are focusing on unraveling mediators of immune evasion, both those that are cancer-cell autonomous as well as those that are tumor microenvironment-mediated, where we focus specifically on the role of the myeloid compartment in modulating T cell responses. We are using a combination of unbiased large-scale CRISPR screens, CaTCH, and novel mouse models followed by the functional workup of candidates and are actively recruiting in this area. Please reach out to us directly.
Identification of novel therapeutic targets in cancers lacking actionable mutations
For many cancers, targeted therapy is not yet an option, because the mechanisms driving tumor progression and specific oncogenic vulnerabilities are unknown. We focused on a set of rare skin cancers, most notably Merkel Cell Carcinoma (MCC), a highly aggressive, neuroendocrine skin cancer. A fraction of MCC patients respond to immunotherapies, but MCC lacks actionable mutations that could be utilized for targeted therapies. Epigenetic regulators, which govern cell fate, provide unexplored therapeutic entry points.
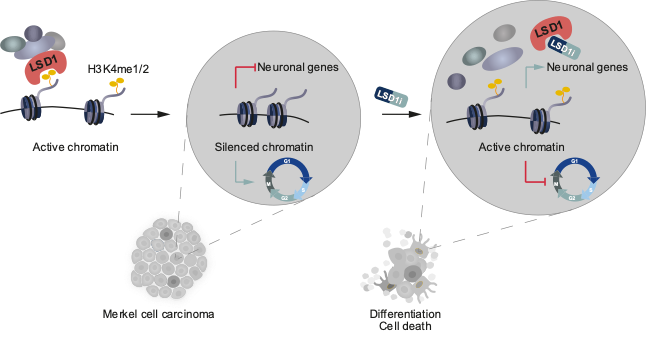
In a manuscript recently published in EMBO Molecular Medicine (Leiendecker, Jung et al., 2020), we identify the Lysine-specific histone demethylase 1A (LSD1, also known as KDM1A) as a new therapeutic target in MCC in vitro and in vivo. We provide detailed molecular insights into the sensitivity and mode of action of LSD1 inhibition in MCC. In ongoing projects, we are expanding our studies to other dependencies we recently identified.
OUR FUNDING







WORD CLOUD FROM OUR PAPERS
